Research Institute of fish Culture and Hydrobiology /RIFCH/
About Laboratory of Molecular, Cellular and Quantitative Genetics
The laboratory engages in both basic and applied research, university education, breeding work and consultancy activities in the field of genetic sources preservation, reproduction and increasing the genetic potential of economically important fish species. Laboratory deals with the research program of centre CENAKVA WP 2 Caviar production technology.
Molecular genetics
We deal in the field of molecular biology with study of genetic and population diversity of economically important fish, crayfish and other crustacean species.
The study is currently focused also on practical application of molecular markers in aquaculture. We utilize in the field of molecular biology:
- Fragmentation analysis of microsatellite markers
PCR-RFLP mitochondrial DNA - Sequencing of mitochondrial and nuclear genes
In the field of fish cellular genetics, we deal especially with studies of evolutionary polyploidy (e.g. in Acipenseridae), of spontaneous polyploidy or induced polyploidy in economically important freshwater fishes (Salmonidae, Cyprinidae, Siluridae), as well as with their biological and breeding impacts. We use cytometric methods for these studies.
- Flow cytometry
- Cell image analyses (2-D and 3-D image cytometry)
Quantitative genetics
The laboratory is involved in the study of fish performance traits. At present, it is focused on growth traits (body length, weight) and yield characteristics (proportions of fillets and processed body) of common carp with the purpose to estimate heritability of these features in the first stage, and in the second stage to register the real feedback to the selection for features with satisfactory heritability. The systematic selection for improvement of genetic potential of growth traits has not been applied for common carp so far, but it is profusely utilized for other fish species.
The field of quantitative genetics, due to fish farming specifics, has to be connected with molecular biology and genetic methods, which enable determination of progeny origin – assigning the progeny to their respective parental pairs within mixed stock. The expansion of the research to the tench or other fish species is expected in the future; and also to MAS (marker assist selection) studies or QTL (Quantitative trait loci) search in common carp.
The field of quantitative genetics is also closely connected to the fish hatchery (Genetic Fisheries Centre of the faculty) with breeding station, or it participates in the program of fish performance traits´ testing at the faculty, as well as in other fishery facilities, and statistically processes collected data.



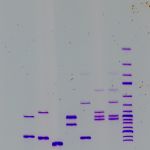
Fragmentation analysis of microsatellite markers
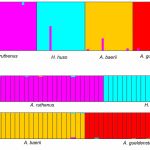
Electroforegram – output of fragmentation analysis of microsatellite loci from CEQ 8000 BeckmanCoulter automated sequencer (left); Result of analysis of microsatellite loci of 4 sturgeon species and interspecific hybrids in STRUCTURE software (right)
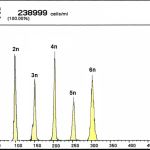
Microsatellites are tandem repeats of short nucleotide patterns distributed within the nuclear genome of eukaryotes. Microsatellites are highly polymorphic DNA markers present as in coding, as in non-coding areas of the genome. Fragmentation analysis of microsatellite markers is used for studying population and genetic diversity of economically important species, such as the common carp (Cyprinus carpio), tench (Tinca tinca), crustaceans (Crustacea, Decapoda), for parentage analyses or for determination of relevance of individuals to populations. Furthermore, this analysis is used for determination of functional ploidy level of acipenserids and, together with current statistic methods, for determination of origin of sturgeons with unusual ploidy level.
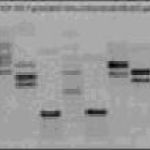
PCR-RFLP of mitochondrial DNA
Electroforetic separation of PCR-RFLP products
Alleles are identified by means of restriction fragment length polymorphism (RFLP) upon presence or absence of a specific restriction site. The amplified genomic DNA (PCR product) is split with respective restriction endonuclease and separated by means of agarose gel electrophoresis. The PCR-RFLP method is used for the study of haplotype diversity of breeds and lines of common carp.
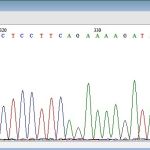
Sequencing of mitochondrial and nuclear genes
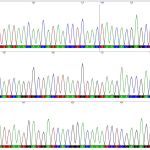
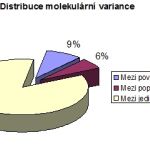
Chromatogram – a graphic output from capillary automated sequencer (part; above), haplotype network constructed in Network software (left); result of analysis of molecular variance (AMOVA; right)
Gene sequencing is used especially for the study of mutual genetic and molecular – biological status of fish and crustacean populations. The analysis itself is focused on genetic characteristics of sturgeon species and of breeds and populations of common carp, tench and sterlet, as well as of crustacean populations in the genus Acanthocyclops (Copepoda: Cyclopidae).
Flow cytometry

Flow cytometry is used especially for routine determination of the ploidy level (haploidy, di-, tri-, tetra-, penta-, hexa-, hepta-, oktaploidy) on the basis of relative volume of DNA in nuclei of somatic fish cells or gametes coloured by DNA-specific dyes. We are working with Partec CCA-I flow cytometer and recently also with CyFlow® Cube 8 multichannel flow cytometer, both from Partec GmBH (Germany). Another possibility is the stipulation of volume and concentration of measured particles.
Image cytometry
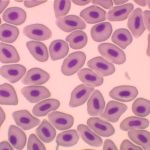
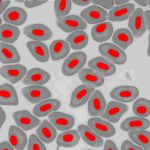
This technique is used besides the basic research of spontaneous, evolutionary and induced polyploidy also in haematology, histology and fish reproduction and within the utilization of genome manipulations in fish breeding.
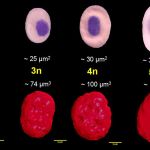
Our most common applications involve analysis of ploidy upon dimensions of fish erythrocyte nuclei (area, perimeter, major axis of nucleus). Comparative methods include analysis of karyotype, quantification of Ag-NOR sites in interphase/metaphase cells, determination of relative DNA content by flow cytometry and determination of genome size (absolute DNA content).
Determination of absolute DNA content in cell nuclei (genome size, C-value) is conducted upon "Feulgen image analysis densitometry, FIA", i.e. application of image analysis for densitometric measurements in cell nuclei in preparations stained with Feulgen reaction. Apart from international standard (chicken, Gallus domesticus; C-value 1.25 pgDNA/nucleus; www.genomesize.com), we use also internal standards of diploid and triploid tench for verification of procedure accuracy (Tinca tinca; C-value 1.01 and 1.55 pgDNA/nucleus). Comparative method is flow cytometric determination of absolute DNA content in PI-stained nuclei.
Computer analysis of macroscopic objects (e.g. eggs, fingerlings, fish, scales etc.) is also used for other biological studies. Partial procedures are applied for verification of origin, performence testing of fish breeds and estimate of breeding value according to the Animal Breeding Act.
We work with image analyzers Cue-2 Image Analyzer (Galai Production Ltd., Israel), Olympus MicroImage version 3.0.1 and 4.0.1 for Windows, CellP (Soft Imaging System GmbH) and Image J (NIH, USA).
Confocal microscopy and 3-D computer image analysis

CellP (Soft Imaging System GmbH) and Image J with numerous plugins are used for processing and measuring of spatial images of cells and cell structures obtained by confocal microscopy. System Olympus LSM Fluoview is coupled to microscope BX50 and enables bi-channel fluorescence microscopy (488 and 568 nm laser) with use of various fluorescent dyes for discrimination of cell structures, scanning of objects in area (XY) and space (XYZ), including scanning in time and computer 3-D reconstructions of scanned objects.
This equipment allows employees and PhD students of the faculty to study relationships between genome size, ploidy level and size of cell and cell nuclei in 3-D space especially in case of fish with higher ploidy level (hexaploidy to dodekaploidy), morphology of blood cells of various fish species from intact populations and from toxicity tests of various water pollutants, morphology and physiology of fish gametes and detailed course of fertilization process.
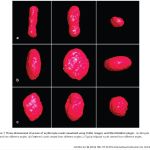
23-D reconstruction of erythrocyte of sterlet (Acipenser ruthenus) stained with Eosine
3-D reconstruction of erythrocyte nuclei of tench (2n and 3n) and of various polyploid sturgeons (4n – 14n)
Staff and contacts